Semantic segmentation using U-Net with PyTorch
Deep learning is here to stay and has revolutionized the way data is analyzed. Furthermore, it is straightforward to get started. Recently, I played around with the fastai library to classify fish species but wanted to go further behind the scenes and dig deeper into PyTorch. As part of another project, I have used a U-Net to perform semantic segmentation of ‘pike’ in images. Training has been done on Google Colab and a local GPU powered workstation excellent for smaller experiments.
The data
Data consists of images of northern pike (Esox lucius). 60 images have been annotated using VIA where regions of ‘pike’ have been delineated using poly-lines. These annotations are used to produce the target masks needed for training. The first part thus consists of loading image and annotation data and convert polygon annotations to images. Furthermore, the original high-resolution images are resized to a common maximum side length (1024 pixels), while maintaining the aspect ratio, to reduce the workload for image IO during training.
Load libraries and data and define paths:
#import libraries and define paths
import os
import sys
import json
import numpy as np
from skimage.io import imread, imsave
from skimage.transform import resize
from skimage.draw import draw
import matplotlib.pyplot as plt
import albumentations as A
import cv2
import random
import torch
from albumentations.pytorch import ToTensorV2
from unet_network import UNet
import time
import copy
data_dir = '../data/'
image_dir = os.path.join(data_dir, "images_pike")
image_small_dir = os.path.join(data_dir, "images_pike_small")
mask_dir = os.path.join(data_dir, "images_pike_masks")
mask_small_dir = os.path.join(data_dir, "images_pike_masks_small")
via_proj = os.path.join(data_dir, "via_project.json")
img_longest_size = 1024
#get VIA annotations from json file
annotations = json.load(open(via_proj))
annotations = list(annotations['_via_img_metadata'].values())
annotations = [a for a in annotations if a['regions']]
#function for reducing high resolution images to have a maximum height or
#width no longer than max_length
def img_max_length(img, max_length, mode):
heigth, width = img.shape[:2]
aspect = width/float(heigth)
if (mode == "img"):
kwargs = {"order": 1, "anti_aliasing": True, "preserve_range": False}
elif (mode == "mask"):
kwargs = {"order": 0, "anti_aliasing": False, "preserve_range": True}
else:
return(1)
if (heigth > max_length or width > max_length):
if(aspect<1):
#landscape orientation - wide image
res = int(aspect * max_length)
img_small = resize(img, (max_length, res), **kwargs)
elif(aspect>1):
#portrait orientation - tall image
res = int(max_length/aspect)
img_small = resize(img, (res, max_length), **kwargs)
elif(aspect == 1):
img_small = resize(img, (max_length, max_length), **kwargs)
else:
return(1)
if (mode == "mask"):
img_small = img_small.astype("uint8")
return(img_small)
else:
return(img)Create image masks from polygons:
#create masks and write to file in both original and reduced size
for a in annotations:
polygons = [r['shape_attributes'] for r in a['regions']]
image_path = os.path.join(image_dir, a['filename'])
image = imread(image_path)
heigth, width = image.shape[:2]
mask = np.zeros([heigth, width], dtype=np.uint8)
rr, cc = draw.polygon(polygons[0]['all_points_y'], polygons[0]['all_points_x'])
mask[rr, cc] = 1
mask_name = a['filename'].replace(".jpg", ".png")
mask_path = os.path.join(mask_dir, mask_name)
imsave(mask_path, mask)
mask_small_path = os.path.join(mask_small_dir, mask_name)
mask_small = img_max_length(mask, img_longest_size, mode = "mask")
imsave(mask_small_path, mask_small)
#create reduced sized versions of high res images
images_raw = list(sorted(os.listdir(os.path.join(data_dir, "images_pike"))))
for i in images_raw:
img_path = os.path.join(image_dir, i)
img = imread(img_path)
img_name = i.replace(".jpg", ".png")
img_small_path = os.path.join(image_small_dir, img_name)
img_small = img_max_length(img, img_longest_size, mode = "img")
imsave(img_small_path, img_small)Setup model and dataset
The model is a U-Net implementation where the input is a 3 channel image and output is a segmentation mask with pixel values from 0-1.
To load the data, we extend the PyTorch Dataset class:
#define dataset for pytorch
class PikeDataset(torch.utils.data.Dataset):
def __init__(self, images_directory, masks_directory, mask_filenames, transform=None):
self.images_directory = images_directory
self.masks_directory = masks_directory
self.mask_filenames = mask_filenames
self.transform = transform
def __len__(self):
return len(self.mask_filenames)
def __getitem__(self, idx):
mask_filename = self.mask_filenames[idx]
image = cv2.imread(os.path.join(self.images_directory, mask_filename))
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
mask = cv2.imread(os.path.join(self.masks_directory, mask_filename),
cv2.IMREAD_UNCHANGED)
mask = np.expand_dims(mask, 2)
mask = mask.astype(np.float32)
if self.transform is not None:
transformed = self.transform(image=image, mask=mask)
image = transformed["image"]
mask = transformed["mask"]
return image, maskTo improve training and generalization, image augmentation is applied. This procedure generates images with random distortions or modifications. The Albumentations library makes this process easy to apply simultaneously for image and mask. Here, transformations are defined for the training set and just resizing and normalization for the validation set:
#define augmentations
train_transform = A.Compose([
A.Resize(400, 400, always_apply=True),
A.RandomCrop(height=200, width = 200, p=0.2),
A.PadIfNeeded(min_height=400, min_width=400, border_mode=cv2.BORDER_CONSTANT,
always_apply=True),
A.VerticalFlip(p=0.2),
A.Blur(p=0.2),
A.RandomRotate90(p=0.2),
A.ShiftScaleRotate(p=0.2, border_mode=cv2.BORDER_CONSTANT),
A.RandomBrightnessContrast(p=0.2),
A.RandomSunFlare(p=0.2, src_radius=200),
A.RandomShadow(p=0.2),
A.RandomFog(p=0.2),
A.Normalize(mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225)),
ToTensorV2(transpose_mask=True)
]
)
val_transform = A.Compose([
A.Resize(400, 400, always_apply=True),
A.Normalize(mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225)),
ToTensorV2(transpose_mask=True)
]
)And finally, the dataset is split into a training (50) and validation set (10):
#split files at random into training and validation datasets
mask_filenames = list(sorted(os.listdir(mask_small_dir)))
random.seed(43)
random.shuffle(mask_filenames)
n_val = 10
val_mask_filenames = mask_filenames[:10]
train_mask_filenames = mask_filenames[10:]
train_dataset = PikeDataset(image_small_dir, mask_small_dir, train_mask_filenames,
transform=train_transform)
val_dataset = PikeDataset(image_small_dir, mask_small_dir, val_mask_filenames,
transform=val_transform)Training the model
Before training, dataloaders are created from the datasets and a few things are defined: a loss function (diceloss), hyperparameters (epochs, learning rate), model instantiated and moved to device (e.g. GPU):
#dice loss function
class DiceLoss(torch.nn.Module):
def __init__(self):
super(DiceLoss, self).__init__()
self.smooth = 1.0
def forward(self, y_pred, y_true):
assert y_pred.size() == y_true.size()
y_pred = y_pred[:, 0].contiguous().view(-1)
y_true = y_true[:, 0].contiguous().view(-1)
intersection = (y_pred * y_true).sum()
dsc = (2.*intersection+ elf.smooth)/(y_pred.sum()+y_true.sum()+self.smooth)
return 1. - dsc
#function to return data loaders
def data_loaders(dataset_train, dataset_valid, bs = 5, workers = 5):
def worker_init(worker_id):
np.random.seed(42 + worker_id)
loader_train = torch.utils.data.DataLoader(
dataset_train,
batch_size=bs,
shuffle=True,
drop_last=True,
num_workers=workers,
worker_init_fn=worker_init,
)
loader_valid = torch.utils.data.DataLoader(
dataset_valid,
batch_size=bs,
drop_last=False,
shuffle=False,
num_workers=workers,
worker_init_fn=worker_init,
)
return {"train": loader_train, "valid": loader_valid}
#define datasets and load network from file
loaders = data_loaders(train_dataset, val_dataset)
device = torch.device("cpu" if not torch.cuda.is_available() else "cuda")
unet = UNet(in_channels=3, out_channels=1, init_features=8)
unet.to(device)
dsc_loss = DiceLoss()
best_validation_dsc = 1.0
lr = 0.001
epochs = 200
optimizer = torch.optim.Adam(unet.parameters(), lr=lr)
best_val_true = []
best_val_pred = []
epoch_loss = {"train": [], "valid": []}Then, the the model can be trained in a couple of minutes (depending on hardware). Best models are saved and training can be monitored from the validation loss:
#training loop
for epoch in range(epochs):
print('-' * 100)
since = time.time()
best_model_wts = copy.deepcopy(unet.state_dict())
for phase in ["train", "valid"]:
if phase == "train":
unet.train()
else:
unet.eval()
epoch_samples = 0
running_loss = 0
for i, data in enumerate(loaders[phase]):
x, y_true = data
x, y_true = x.to(device), y_true.to(device)
epoch_samples += x.size(0)
optimizer.zero_grad()
with torch.set_grad_enabled(phase == "train"):
y_pred = unet(x)
loss = dsc_loss(y_pred, y_true)
running_loss += loss.item()
if phase == "train":
loss.backward()
optimizer.step()
epoch_phase_loss = running_loss/epoch_samples
epoch_loss[phase].append(epoch_phase_loss)
if phase == "valid" and epoch_phase_loss < best_validation_dsc:
best_validation_dsc = epoch_phase_loss
print("Saving best model")
best_model_wts = copy.deepcopy(unet.state_dict())
best_val_true = y_true.detach().cpu().numpy()
best_val_pred = y_pred.detach().cpu().numpy()
time_elapsed = time.time() - since
print("Epoch {}/{}: Train loss = {:4f} --- Valid loss = {:4f} --- Time: {:.0f}m {:.0f}s".format(epoch + 1, epochs, epoch_loss["train"][epoch], epoch_loss["valid"][epoch], time_elapsed // 60, time_elapsed % 60))
print("Best validation loss: {:4f}".format(best_validation_dsc))
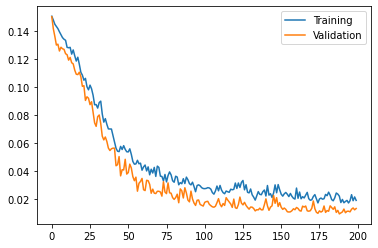
torch.save(best_model_wts, os.path.join(data_dir, "unet.pt"))Training and validation run from 200 epochs:
Using the model
After training, the model weights are saved to a file. These can be re-loaded and used with the network for producing predictions for new samples:
#load weights to network
weights_path = data_dir + "unet.pt"
device = "cpu"
unet = UNet(in_channels=3, out_channels=1, init_features=8)
unet.to(device)
unet.load_state_dict(torch.load(weights_path, map_location=device))Then, a couple of utility functions are loaded to resize the image to fit the network, apply predictions and putting it all together:
#define augmentations
inference_transform = A.Compose([
A.Resize(400, 400, always_apply=True),
A.Normalize(mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225)),
ToTensorV2()
])
#define function for predictions
def predict(model, img, device):
model.eval()
with torch.no_grad():
images = img.to(device)
output = model(images)
predicted_masks = (output.squeeze() >= 0.5).float().cpu().numpy()
return(predicted_masks)
#define function to load image and output mask
def get_mask(img_path):
image = cv2.imread(img_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
original_height, original_width = tuple(image.shape[:2])
image_trans = inference_transform(image = image)
image_trans = image_trans["image"]
image_trans = image_trans.unsqueeze(0)
image_mask = predict(unet, image_trans, device)
image_mask = F.resize(image_mask, original_height, original_width,
interpolation=cv2.INTER_NEAREST)
return(image_mask)Finally. the predictions for a new image, which has not been used for training:
#image example
example_path = "159_R_1.png"
image = cv2.imread(example_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
mask = get_mask(example_path)
masked_img = image*np.expand_dims(mask, 2).astype("uint8")
#plot the image, mask and multiplied together
fig, (ax1, ax2, ax3) = plt.subplots(3)
ax1.imshow(image)
ax2.imshow(mask)
ax3.imshow(masked_img) The result after training a U-Net to perform semantic segmentation of ‘pike’. The top is the new and unseen (by the model) image, the middle is the predicted mask region and the bottom is the two multiplied to apply the mask
The result after training a U-Net to perform semantic segmentation of ‘pike’. The top is the new and unseen (by the model) image, the middle is the predicted mask region and the bottom is the two multiplied to apply the mask
Conclusions
- Image-augmentation improves training and Albumentations makes this easy
- Images can quickly be annotated using VIA
- U-Net does not require large datasets and performs well
- Getting ‘under the hood’ using PyTorch directly is a learn full experiences